The enemy within: Regulating the expression of mutagenic DNA
John Karijolich, Ph.D., Discovery Grant 2017
Annually, over 1 million Americans are diagnosed with cancer, and a further approximate 600,000 will lose their battle. At its heart, cancer results from a loss of the controls that tell a cell when to stop growing. In the human body, there are trillions of cells with various functions. These cells grow, divide, and multiply to help the body function properly. When cells become old or damaged they die, and new cells replace them. Cancer develops when the body’s normal control mechanisms stop working. Old cells do not die and they grow out of control, forming new, abnormal cells.
The human genome carries the blueprint for all our working parts. It contains an estimated 20,000 to 25,000 protein-coding genes. Genes are “expressed” in an ordered fashion: the code is our DNA, which drives the formation of RNA, which is read to make proteins. These proteins are what you see when you look at your father, mother, child—they compose our skin, hair, and even that ingrown toenail you’ve been meaning to get looked at. Some of these 20,000 – 25,000 genes, when their expression is permanently altered, can lead to the development of cancer. Understanding the regulation of gene expression is thus inherently relevant to all cancers.
While much of biomedical research has been focused on investigating the function of proteincoding genes, strikingly they account for only about 2% of our genome. The other 98%, which has been referred to as the “dark matter” of our genome, is where research in the Karijolich laboratory is focused. Within the dark matter lie relics of our past, genetic fossils of viruses that infected our ancestors many millennia ago, as well as evolution attempts at new gene construction. Collectively, we refer to these elements as “retrotransposons”, and they account for approximately 45% of our total genome.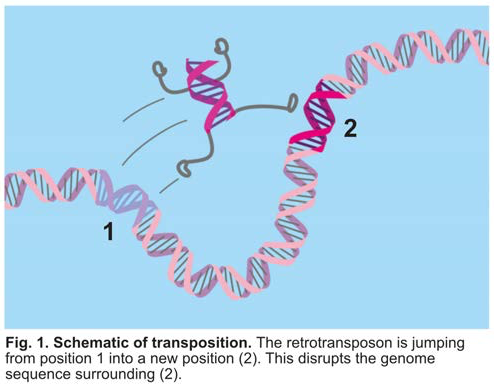
Retrotransposons are not expressed in healthy cells; however, for reasons that are not understood, occasionally retrotransposons can be expressed. What makes retrotransposon expression so potentially harmful is the fact that some of them are able to spread (transposon) throughout the genome. The spreading process, termed transposition, can cause gene mutations because the retrotransposon sequence is being inserted into a place in which it was previously not present (Fig. 1). In fact, previous research has found that transposition into tumor suppressor genes can cause cancer. Furthermore, continuous spreading of retrotransposons within cancerous cells can result in additional mutations within the genome, making it more likely that they do not respond to therapy and increasing the likelihood of metastasis. Thus, defining how retrotransposons escape the control mechanisms that should silence their expression will be key to preventing their escape as well as their sustained expression in cancerous cells.
This proposal aims to uncover the cellular mechanisms that regulate retrotransposon expression. We are interested in how DNA sequence elements near retrotransposons are altered to allow their expression. In addition, cellular DNA is wrapped in a complex of proteins called chromatin, and we are interested in whether alterations of chromatin facilitate retrotransposon expression. Results from these studies will be highly applicable to a range of cancers, including some of the most common cancers today, such as breast, lung, and colon, all which show robust retrotransposon expression. The therapeutic silencing of retrotransposon expression may provide a means to limit further genomic damage in cancerous cells.


